My favorite molecule
A dozen Stanford Medicine researchers explain what piques their interest when it comes to molecules

For kids on a playground, it’s a common refrain between new friends: “What’s your favorite color? What’s your favorite animal?”
For young adults, they make good icebreakers on dates: “What’s your favorite place in the world? What’s your favorite pizza topping?”
A person’s favorites often cut to the punch of their personality, likes and preferences — although, admittedly, a favorite book or hobby might say more about someone than their favorite dinosaur.
In the professional world, favorites rarely come up, but surely scientists have them, whether they’re favorite conferences, journals, papers or experiments.
For this issue, we asked 12 Stanford Medicine researchers to tell us about their favorite molecules.
It turned out to be a good way to give insight into their research: Every scientist — perhaps unsurprisingly — named a molecule that they actively study.
In most cases, the potential impact of a molecule on human health topped the list of reasons a researcher calls it a favorite.
But for some scientists, they consider a molecule their favorite because it is understudied, has multiple roles in the body, has a neat three-dimensional structure or has been key to their professional success.
Manuel Amieva, MD, PhD
Professor of Pediatrics and of Microbiology and Immunology
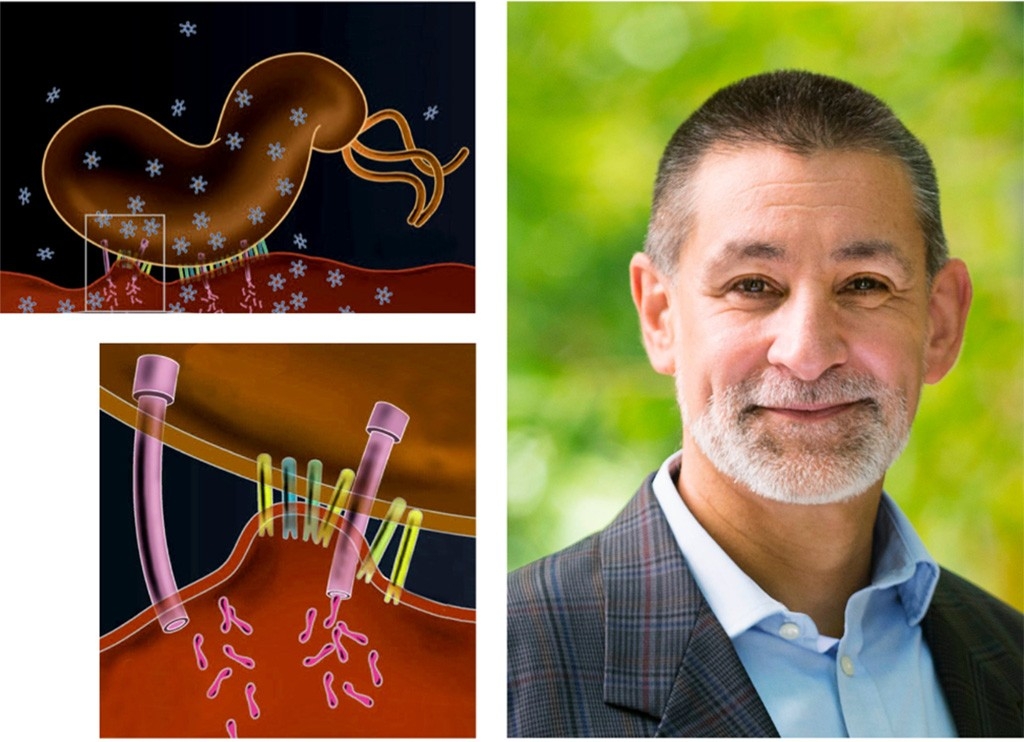
Cag bacterial type IV secretion system
When Helicobacter pylori bacteria slide down someone’s esophagus into the stomach (and they do this often, having infected about half the world’s population), they are armed for trouble.
The microbes are coated with miniscule needles — called the bacterial type IV secretion system — that poke into human cells to inject proteins.
“I find it amazing that bacteria evolved to have these tricky little nano-syringes,” said Manuel Amieva.
In H. pylori, the type IV secretion system is quite complex, with about a dozen different protein subunits, but it has just one known job: delivering a protein known as CagA, which can boost people’s risk of ulcers and stomach cancer. For the past two decades, Amieva has studied CagA — how it can contribute to cancer risk and why H. pylori has the protein in the first place.
The secretion system and its payload, which Amieva ranks as his favorite molecular structure, are an example of how bacteria have evolved sophisticated communications systems with their host, he said.

Alka Das, PhD
Postdoctoral Scholar in Molecular and Cellular Physiology
MEC-4
In the Caenorhabditis elegans roundworm used frequently in biology labs, the protein MEC-4 acts like a tiny tripwire. The doughnut-shaped MEC-4 spans the membrane of some C. elegans cells. When a predator pokes, prods or squeezes the worm, MEC-4 senses the pressure and opens its central channel, letting charged molecules flow through.
“When worms don’t have MEC-4, they actually can’t sense any gentle touch,” said Alka Das, who studies the protein. “It’s really fascinating to me that these proteins can translate a physical force into a biological response.”
While scientists have characterized other touch-sensitive ion channels, MEC-4 has a drastically different sequence and is finicky in the lab, Das said. She is still trying to determine its three-dimensional structure as well as what other proteins it interacts with.
Michael Fischbach, PhD
Associate Professor of Bioengineering and of Medicine
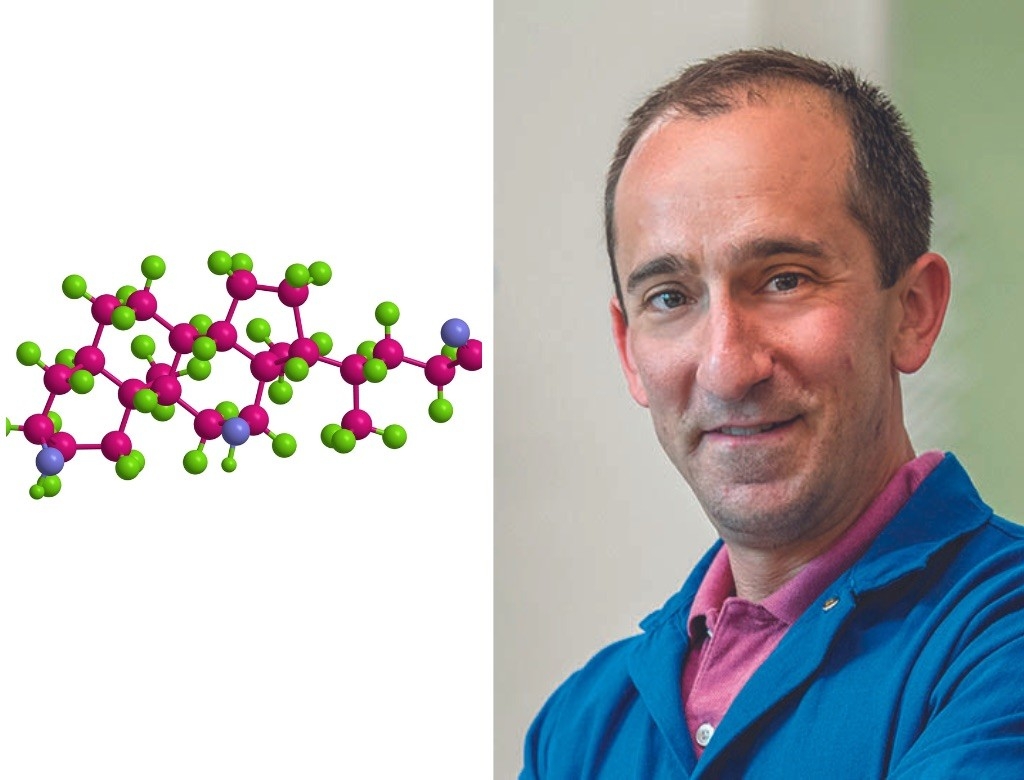
Deoxycholic acid
For the past five years, Michael Fischbach has been enamored of a molecule that, he said, “basically makes up the exhaust fumes of bacteria in the gut.” Our intestines, however, put this bacterial waste — deoxycholic acid — to good use, using it to absorb fats in our intestines.
“I’m fascinated by this fact that there are hundreds of molecules in circulation in our bodies that weren’t made by us but impact our biology,” said Fischbach.
Deoxycholic acid not only digests fatty foods better than cholic acid — the version of the molecule that our own bodies make — but also binds to receptors in the intestines, having far-reaching effects on metabolism and immunity.
Roger Kornberg, PhD
Mrs. George A. Winzer Professor in Medicine
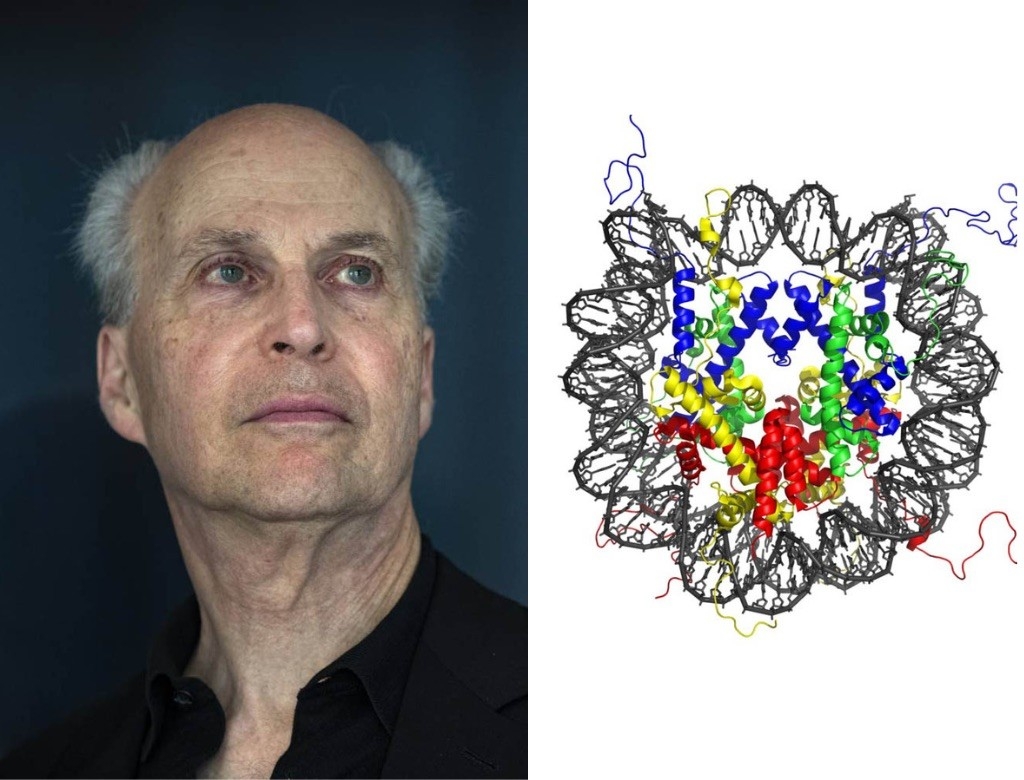
The Nucleosome
A strand of DNA, yanked out of a cell, is roughly as long as an average adult is tall. In 1974, Roger Kornberg discovered how nucleosomes — clusters of proteins bound to DNA like beads along a string — package this lanky genetic material into tightly folded structures inside cells.
“The nucleosome was probably the most important discovery of my career,” said Kornberg, whose career includes a Nobel Prize in Chemistry. “Working alone, as a very young scientist, I conceived of the solution of this DNA packaging problem that had been studied for decades.”
In the years since, Kornberg and his wife and longtime collaborator, Yahli Lorch, PhD, have revealed how nucleosomes are more than just physical spools; the molecular complexes control which genes are expressed in which cells in the body. Genetic material wound around nucleosomes can’t be accessed by the cellular machinery that reads DNA, so the packaging of nucleosomes determines which genes a cell is using.
Jonathan Long, PhD
Assistant Professor of Pathology
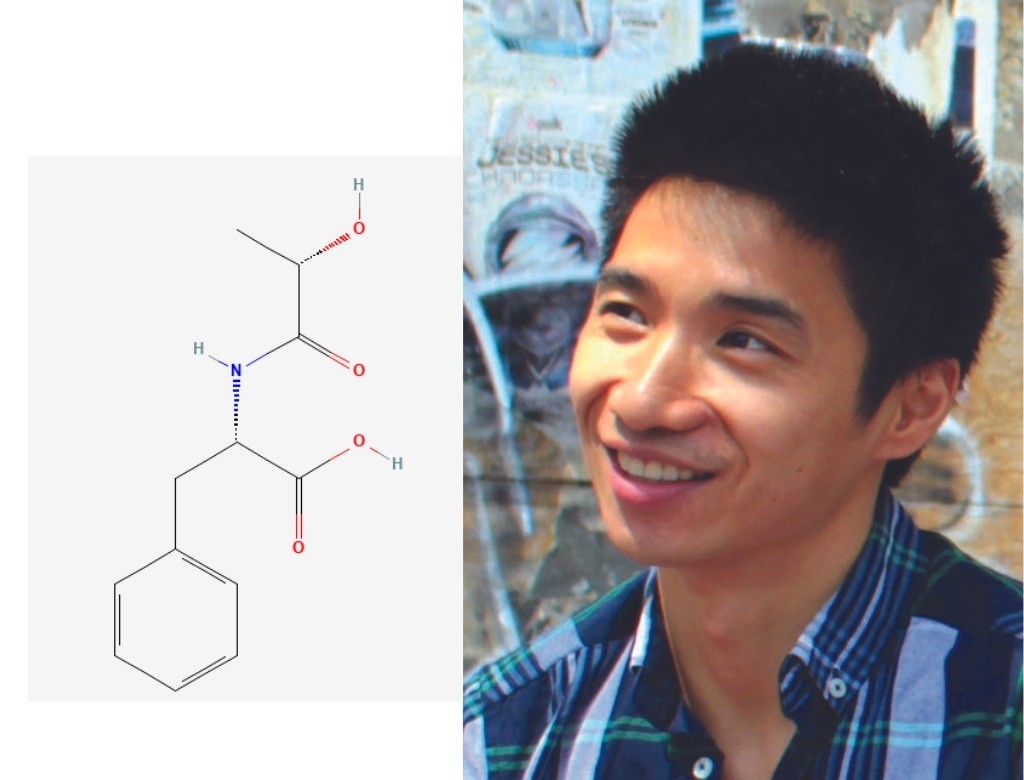
Lac-Phe
If you’ve ever felt queasy after a long run or lost your appetite for breakfast after a morning spin session, then you have firsthand experience with Jonathan Long’s favorite molecule: a chemical known as Lac-Phe.
Long’s lab group recently discovered that this tiny signaling molecule shuts down hunger signals in the brain after strenuous exercise. When you exercise, lactic acid builds up in your muscles, creating several bioactive byproducts, one of which is Lac-Phe.
Long’s team members think that during and after a workout session, the molecule races from the muscles to the brain and acts on neurons to turn down the dial on hunger, though the researchers haven’t pinpointed just how this happens.
“What’s neat is that Lac-Phe is present in every animal that moves, meaning it’s extremely conserved and ancient,” said Long. “Understanding it better might help us capture the benefits of exercise to treat all kinds of things, from osteoporosis to obesity.”
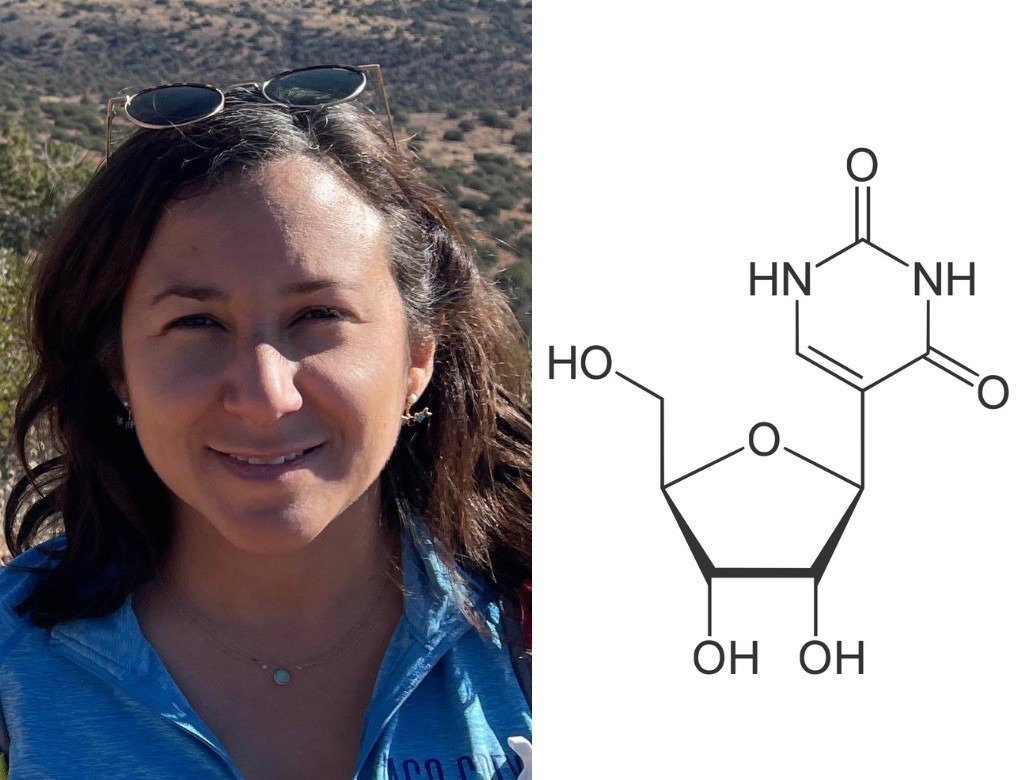
Nicole Martinez, PhD
Assistant Professor of Chemical and Systems Biology and of Developmental Biology
Pseudouridine
Nicole Martinez’s favorite molecule is like a code within a code. Imagine you were reading an instruction manual and every time you saw a backwards R, you’d have to pull the manual apart, assemble it in a new way and follow new instructions. Those Яs are kind of like pseudouridine — a modified, rotated version of the RNA building block called uridine.
When an RNA molecule contains pseudouridine in place of uridine, it can change how the molecule is processed, how it functions, how stable it is and even whether the immune system can recognize it as RNA. But researchers are stymied by the details on how and why; Martinez is trying to break the code.
“I’m just fascinated by this idea that in addition to the basic sequence of RNA, there’s an additional layer that can change the meaning of RNA ’s message,” said Martinez.

Daria Mochly-Rosen, PhD
Professor of Chemical and Systems Biology and the George D. Smith Professor in Translational Medicine
Drp1
As Daria Mochly-Rosen describes her favorite protein, Drp1, her arms rise in front of her body, elbows swinging one way and then another to show how the hinged protein moves.
“I’m a protein chemist and I just love to look at how proteins move,” she said. “Drp1 is a fun little machine.”
Inside cells, Drp1 molecules arrange themselves, arm over arm, around mitochondria — organelles that, among other things, generate energy. When the Drp1 ring tightens, it pinches a mitochondrion in two. But Drp1 has different partners for this dance. When it binds one protein, mitochondrial division progresses at a normal pace; when it binds another, mitochondria divide too much and too frequently — potentially contributing to neurodegeneration.
Mochy-Rosen’s lab has developed a drug that keeps Drp1 from binding to the more toxic dance partner while still allowing its normal function. “If we learn how to control Drp1, we can improve mitochondrial function, which is essential to treating many pathologies including neurodegenerative diseases,” she said.
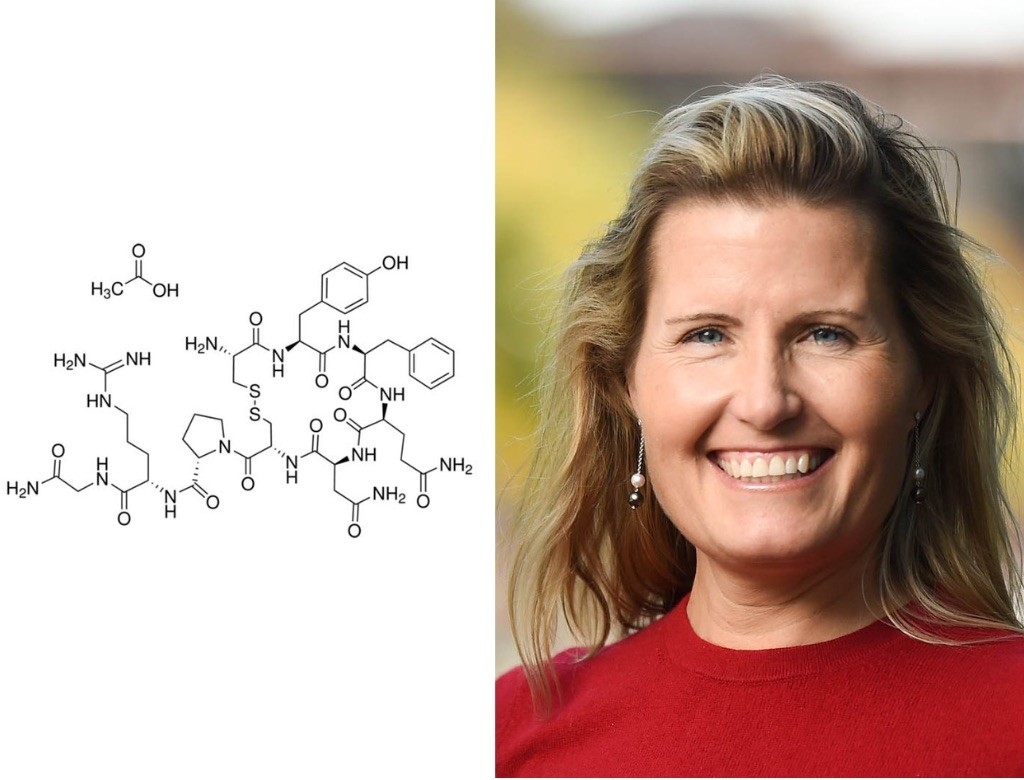
Karen Parker, PhD
Associate Professor of Psychiatry and Behavioral Sciences
Vasopressin
When it comes to molecules, Karen Parker has a soft spot for the underdog. “For many years, vasopressin has been in the shadow of oxytocin, which has gotten all of the scientific attention,” she said. “That’s part of why I like it so much; it’s really under-researched.”
Oxytocin is often called “the love hormone” because its activity in the brain is associated with attachment, trust and social interaction. But Parker thinks that vasopressin is just as important; she discovered that people with social impairment disorders have lower than usual amounts of vasopressin — but not oxytocin — in their spinal fluid. She and her colleagues can even predict which newborns will develop autism based on these vasopressin levels, and she has shown that giving vasopressin to children with autism boosts their social abilities and diminishes anxiety.
“Since we’ve been publishing papers on vasopressin, I’ve seen an uptick in interest,” said Parker. “I hope it continues to get more love.”
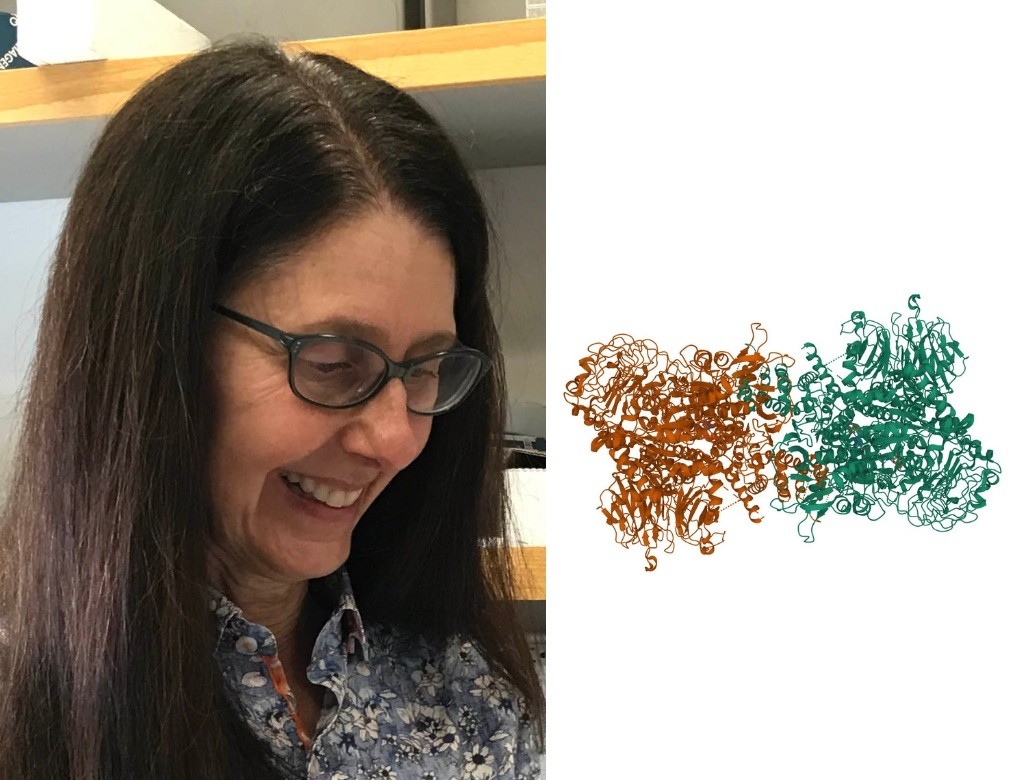
Suzanne Pfeffer, PhD
Professor of Biochemistry and the Emma Pfeiffer Merner Professor in Medical Sciences
LRRK2
When Suzanne Pfeffer learned of LRRK2, she was immediately captivated; LRRK2 is the most commonly mutated protein in inherited Parkinson’s disease, but researchers didn’t understand why.
During the past five years, Pfeffer’s lab group has pieced together much of the mystery, revealing how mutations in LRRK2 make a small set of brain cells lose their primary cilia — miniscule, molecular antennae that nearly every human cell uses to communicate. Without primary cilia, the cluster of neurons in the brain can’t receive vital stress signals sent by neighboring cells, so they don’t send back necessary protective factors. The sets of neurons on both sides of the interaction begin to die.
Some pharmaceutical companies are testing drugs that quell the activity of LRRK2 in patients with Parkinson’s disease. But Pfeffer isn’t moving on from her favorite molecule yet; she thinks it has many important secrets that are yet to be discovered.
“We still don’t know why this one group of cells in the brain is so uniquely sensitive to LRRK2 mutations, considering LRRK2 is found throughout the rest of the brain and body too,” she said.

Kathleen Ruppel, MD, PhDS
enior Scientist in Biochemistry
Myosin
In 1987, Kathleen Ruppel was a first-year MD/PhD student sitting in a Stanford Medicine lecture hall when biochemistry and structural biology professor James Spudich, PhD, showed the class a video of his favorite molecule as it moved fluorescent filaments across a microscope slide. It was myosin — a tiny motor that powers movements within cells.
“I was hooked on myosin pretty much right away,” said Ruppel.
Myosin has remained Ruppel’s favorite molecule, and — now a pediatric cardiologist — she has teamed up with Spudich to study its array of functions in the human body, especially the heart. Versions of myosin not only power muscles, from biceps to the beating heart, but also play roles in the smaller-scale movements of materials in and out of cells.
Ruppel said the fact that each type of myosin molecule has its own quirk keeps the research interesting; some walk hand over hand like orangutans hanging from a branch while others stroke in synchrony like oars in a boat.

Peter Santa Maria, MD, PhD
Associate Professor of Otolaryngology – Head and Neck Surgery
Heparin-binding EGF-like growth factor
Peter Santa Maria’s molecule of choice is an earful, both literally and figuratively. His lab discovered in 2014 that heparin-binding EGF-like growth factor, or HB-EGF for short, can regenerate a damaged eardrum. While some people’s eardrums manage to repair themselves after a rupture, others never heal on their own. These unhealed ruptures — especially common in developing nations — can cause hearing loss and recurrent ear problems.
Santa Maria’s lab showed that, in mice, HB-EGF can coax chronic eardrum perforations to close. The pharmaceutical company Astrellas Pharma Inc. licensed the therapy, from a startup that Santa Maria co-founded, and is now enrolling patients in an early-phase study to test how well HB-EGF works in humans with chronically ruptured eardrums.
“This is the molecule that I’ve had the most joy with,” said Santa Maria. “If it keeps performing well in clinical trials, I think it will be my favorite for life.”
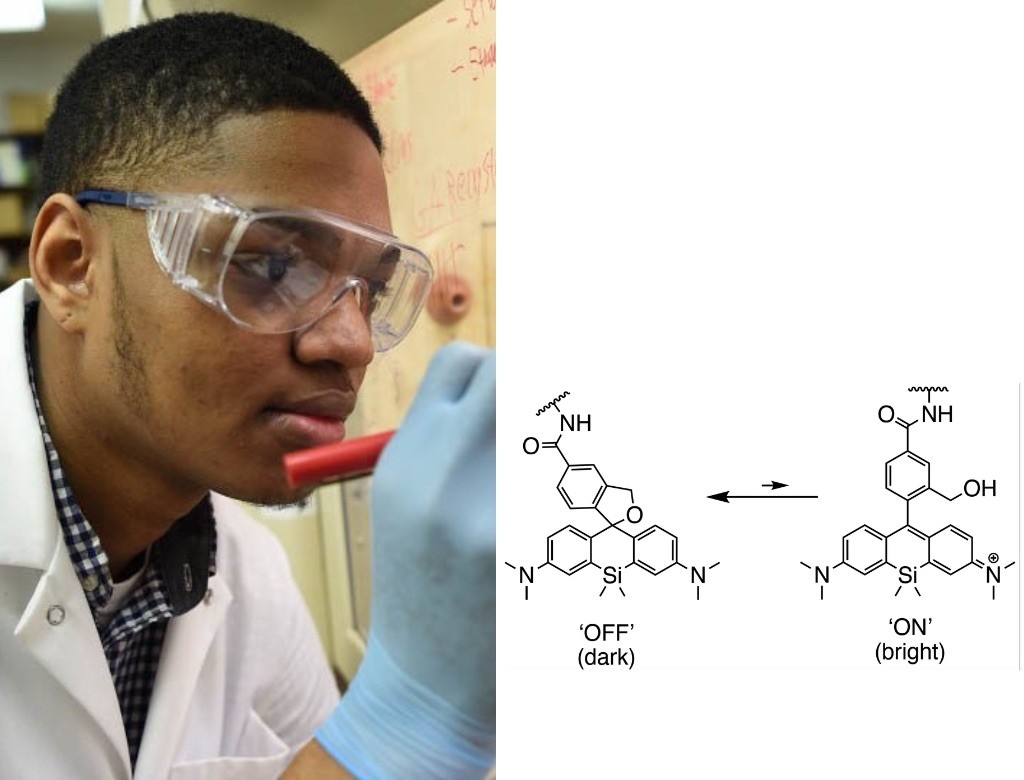
Jonathan Tyson, PhD
Postdoctoral Scholar in Bioengineering
Hydroxymethyl silicon rhodamine
Trying to peer inside a living cell and keep track of all the components is like trying to hear a conversation in a noisy cafeteria, said Jonathan Tyson. Everything blurs together. The fluorescent molecule hydroxymethyl silicon rhodamine, or HMSiR, changes that; unlike most fluorescent tags that remain steadily on, it blinks on and off, illuminating at any given time just a small percentage of the molecules that it is attached to.
“HMSiR makes every molecule speak one at a time, with decorum,” said Tyson. “Suddenly you can understand what they’re all saying.”
Tyson relied on HMSiR to carry out his graduate work, revealing the minute details of cellular organelles and their movements and shape-shifting over long periods of time.
He also designed other, similar molecules from scratch to allow more precise, nano-scale microscopy. Now, he’s studying how related “blinking” molecules might be useful in building molecular devices on and in cells rather than simply illuminating cellular components.

