Dawn of the ‘surprisosome’
Ribosomes, the gene’s translators, upend the central dogma of genetics

A highly trained orchestra can play any musical score set in front of it. But what if a simple swap of a flute with a piccolo limited the set list strictly to military marches, or the switch of a French horn for a clarinet meant listeners would be swaying in their seats to waltzes all evening?
It would be a whole different concert experience — and probably not one you signed up for when you bought your ticket.
“This is very unexpected,” said Stanford School of Medicine associate professor of genetics Maria Barna, PhD.
Barna was, of course, not talking about orchestras, but about ribosomes — the cellular factories that turn genetic messages from our DNA into proteins responsible for nearly every function in our cells. They are the ultimate all-purpose machines; every protein in your body, in anybody’s body, in fact, was birthed by one of the multitude of ribosomes bobbing in each cell’s cytoplasmic soup.
They are also almost bafflingly ancient. The ribosome’s core structure arose more than 3 billion years ago and is shared by every living organism. As a result, many researchers have believed for decades that ribosomes were mostly identical and largely indiscriminate; little mechanical pianos mindlessly reading any and all nearby genetic messages and obediently spewing out the encoded proteins into the cytoplasm like melodies in a concert hall.
Recently, though, Barna’s work has illuminated a secret life for ribosomes that upends our fundamental understanding of how the genetic code gives rise to the cells and tissues in our bodies. Like that fickle orchestra, Barna has found, ribosomes are unexpectedly picky: Some prefer to latch primarily onto messages important to how a cell uses nutrients (polkas!), others to messages that dictate how and when the cell divides and develops (tangos!).
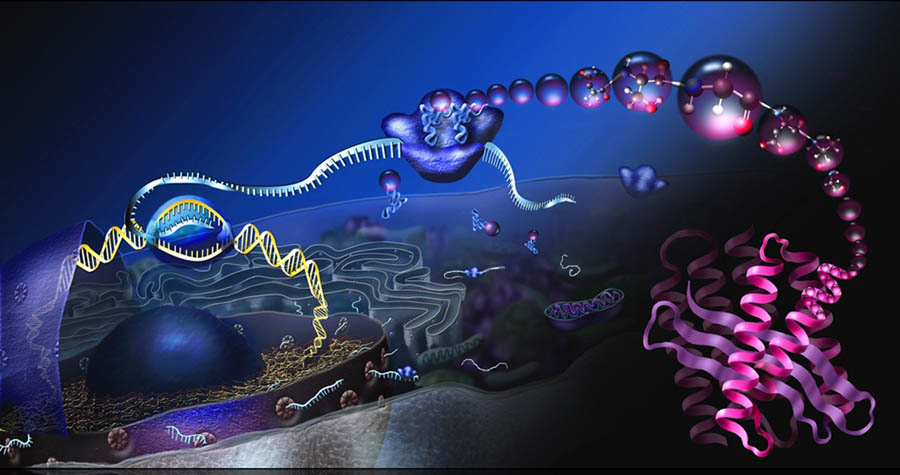
These predilections mean that ribosomes act as de facto gatekeepers, controlling in broad sweeps which genres of messages are made into proteins and freeing cells from micromanaging the relative levels of thousands of individual genes. In doing so, these ubiquitous factories play critical roles in cell fate and function and even control the earliest steps of embryonic development and how and where body parts are formed.
“This is a completely unexpected way to control body plan formation, and an entirely new frontier of study,” Barna said. “We’re learning that evolution bakes in layers of genetic regulation whenever and wherever possible to allow cells to quickly respond to changing environmental conditions or developmental demands.”
“This is pretty spectacular data,” said Robert Schneider, PhD, the Albert Sabin Professor of Molecular Pathogenesis at NYU Grossman School of Medicine. “Maria has shown clearly that cells, which are smarter than we are, have ways to alter the composition of ribosomes in ways that impact the kinds of proteins they make.”
But not everyone has been happy with the ribosomes’ move from the wings to center stage. “Some scientists studying hardcore ribosome structure — how ribosomal proteins interact at a molecular or atomic level — and who favor a reductionist approach to ribosomal biology view these findings as controversial,” Barna said. “However, we’ve shown that ribosomes are dynamic, and each cell can have its own constellation of ribosomes. This has critical implications for biomedical research.”
“We’ve shown that ribosomes are dynamic, and each cell can have its own constellation of ribosomes.”
Maria Barna, PhD, associate professor of genetics
High school biology students learn that genes are found on long stretches of DNA that make up chromosomes. These genes are selectively copied, or transcribed, into short stretches of messenger RNA that carry the assembly instructions for specific proteins.
Because only genes that are transcribed into RNA can become proteins, scientists have focused for decades on the process of transcription as the key arbiter of when and where and which proteins are made. They’ve identified hundreds of specialized proteins called transcription factors that can dial the transcription of specific genes up or down based on other nearby DNA sequences. Other proteins act more globally, controlling the transcription factors’ access to longer stretches of DNA by either keeping it tightly packed or allowing it to unspool for rapid transcription.
These layers of control, or regulation, ensure that cells have enough of the right kinds of proteins at the appropriate times to carry out their specific tasks — a muscle cell might need plenty of energy-generating factories called mitochondria, for example, while a rapidly dividing cell might need to ramp up its production of molecules involved in the cytoskeleton that helps each new daughter cell hold its shape. But researchers, including Barna, have uncovered clues that the control of transcription isn’t the whole story.
“After the birth of messenger RNA, there are additional layers of gene regulation,” Barna said. “But these downstream steps are much less well understood. What we’ve come to realize is that the levels of transcription of a particular gene are a poor proxy for the amount of protein encoded by that gene that is actually made in the cell.”
Enter the ribosome
Every living cell has thousands or even millions of ribosomes crammed into their cytoplasm waiting to translate the instructions in messenger RNA molecules into proteins. These highly specialized machines are made up of 79 core proteins, clustered around a heart of four RNA molecules that NASA scientists recently confirmed evolved before life itself. Together, the proteins and RNA form two subunits, one that binds to and reads, or translates, the messenger RNA molecule and another that recruits the amino acids called for by the RNA blueprint and joins them together like popcorn on a string.
Ribosomes were first identified in 1955 by George Palade, who became intrigued by electron microscopic images that showed a profusion of tiny speckles peppering a cross section of rat pancreas. He was awarded the Nobel Prize in physiology or medicine in 1974 for his discovery that the ribosomes were solely responsible for protein synthesis in cells.
“Some people studying the control of translation pushed the idea that ribosomes are just big, lumbering machines that all translate every messenger RNA the same way, and that has stuck for decades.”
Robert Schneider, PhD, of the NYU Grossman School of Medicine
At first blush, all ribosomes seemed to be basically the same. Though some early hints emerged that ribosomal structure and composition may differ among species, that idea fell out of favor during the 1960s and ’70s as scientists pushed to understand the genetic code and its expression in the broadest possible terms.
“Biologists tend to want to make things simple,” said Schneider, “even though nature is not simple. Some people studying the control of translation pushed the idea that ribosomes are just big, lumbering machines that all translate every messenger RNA the same way, and that has stuck for decades.”
But one conundrum was difficult to explain away: Several human diseases that are caused by mutations in ribosomal proteins, or in the proteins that direct the formation of intact ribosomes from their component proteins, manifest only in specific tissues or cells of the body. Diamond-Blackfan anemia, for example, hobbles cells in the bone marrow that make red blood cells and can also lead to skeletal defects such as a small jaw or a cleft palate. In contrast, North American Indian childhood cirrhosis specifically damages the liver. If ribosomes function identically in all tissues and cells, why were the disorders caused by the mutations so location specific?
Tails of two mice
It was 1946. And mouse #289, a male, was about to meet his sister, mouse #291 — courtesy of the breeding program devised by researchers from the National Cancer Institute in Bethesda, Maryland. Breeding brothers to sisters keeps mouse lineages with genetic traits that scientists want to study pure from the influence of genes from other, unrelated mice.
This lineage, or strain, was known as strain C. And for 62 previous generations, these brother and sister matings had gone off smoothly. But this time there was a hitch. Or, rather, a kink: One of the resulting 18 offspring had an unusually short, kinked tail. And 3 of 26 pups from a subsequent mating between #289 and another sister, #292, also sported the strange appendage. Normally, a laboratory mouse’s tail is about 4 inches long; the tails of these mutant mice ranged from a barely noticeable stub to around 1.5 inches.
The mice had another quirk: Many of them had a variety of skeletal abnormalities, including an extra set of ribs that replaced cervical vertebrae. “Their entire skeletal system is jumbled up,” Barna said. “It was very rare to see so many homeotic, or developmental, changes in a single mouse strain.”
The researchers were intrigued. They named the new mutant mouse strain “tail-short,” and embarked on a breeding program to learn more about the gene that might cause such mutations. But it would be decades before the root cause was discovered, by Barna, in 2011.
Ribosome at work: This video shows an mRNA strand moving through the ribosome as tRNA molecules carry in amino acids to build a protein according to the mRNA code. (Courtesy of Maria Barna)
As an undergraduate at New York University, Barna majored in anthropology. But when she needed to fulfill a science requirement, she ended up in a graduate level course in immunology taught by NYU viral immunologist Carol Reiss, PhD. Soon, Barna was working in Reiss’ laboratory, and by graduation she had published several peer-reviewed papers on viruses that infect the nervous system of mice.
Barna took some time off after her degree to work as a technician in a cancer genetics lab at Memorial Sloan Kettering. When she bred a mouse with strange looking limbs while investigating genes involved in leukemia, she sought out Lee Niswander, PhD, at Sloan Kettering for help. “I became really interested in these uncharacterized mouse mutants, and she was an expert in embryonic development,” Barna said.
One of those mutants was the tail-short mouse identified decades earlier. Barna, who leapfrogged directly from graduate school at Cornell University to an independent researcher with her own lab at UC San Francisco, showed in 2011 that the tail-short mice owed their distinctive skeletal structure to a defect in a ribosomal protein called Rpl38.
“This result was a huge surprise,” Barna said. “We expected to find a master regulator of gene packaging, or a transcription factor that influences gene expression levels. Instead, we found the first link between a ribosomal protein and the fundamental mammalian body plan.”
At UC San Francisco, Barna benefited from a close association with developmental biologist Gail Martin, PhD, who was one of the first people to identify and isolate embryonic stem cells from mice. Their joint lab meetings helped Barna puzzle through her unconventional results.
Rpl38, they learned, is necessary to help the ribosome latch onto messenger RNAs that encode a group of proteins that regulate the formation of the skeleton in mammals — ensuring that all the legs, arms and vertebrae end up in the right place at the appropriate time. The finding was the first to suggest that the presence or absence of specific ribosomal proteins might confer an unexpected selectivity in ribosomal function.
“We began to realize this could have critical implications for our understanding about how genes are regulated within the cell.”
Maria Barna
They also found that the genes for Rpl38 and several other ribosomal proteins vary considerably in their expression among tissue types and developmental stages — suggesting the composition of ribosomes is likely to be much more modular and dynamic than previously expected.
“We began to realize this could have critical implications for our understanding about how genes are regulated within the cell,” Barna said.
In 2012, Barna moved to the Stanford School of Medicine to continue her research. “When I started at Stanford I took a bold approach,” she said. “We wanted to try to answer the question as to whether all ribosomes were the same, or if they did in fact vary.”
Initially they looked at just 15 of 80 ribosomal proteins, using a special technique that compared the ratios of the proteins in embryonic stem cells. “We started to see that not all the proteins were present in all ribosomes,” Barna said. “So there must be some level of regulation going on. Each cell can have its own constellation of ribosomes, and a new level of control of gene expression.”
They also found that ribosomes collect a variety of ribosome-associated proteins, or RAPs, that dangle off their outer shells like ornaments on a Christmas tree. Some of these RAPs are also components of key molecular pathways governing the cell cycle, energy metabolism and signaling between cells.
The RAPs, they learned, further fine-tune the ribosome’s preference for which messages they choose to translate — helping them to select from a vast library of messenger RNA floating in the cell’s cytoplasm. To extend the orchestra metaphor, perhaps one cues up square dances and classical scores while another prefers disco and swing. Each choice results in a specialized panel of proteins that can direct cell fate.
The ribosome’s importance in embryonic development was cemented in 2022, when Barna and her lab showed that mutations in another ribosomal protein, Rpl10A, result in mouse embryos that one-up the tail-short mouse of the 1940s.
Mice with Rpl10A completely lack not just a tail, but a chunk of their body. “It is a striking body structure,” Barna said. “It’s as if a guillotine had chopped off all tissue after their hind limb, but the rest of the organs were completely normal.”
Further research showed that the mutations affected the ability of the embryos to make mesoderm — one of three primary tissue types, or germ layers, from which all other body parts arise. Rpl10A, they found, promotes the ability of ribosomes to translate messenger RNA molecules that encode members of the Wnt family of proteins — another group notorious for their roles in tissue patterning. Rpl10A also waxes and wanes dramatically in its abundance in ribosomes during early embryonic development in a pattern mirroring mesoderm development.
“This is likely to be the tip of the iceberg,” said Naomi Genuth, PhD, a former graduate student in Barna’s lab and the first author of a 2022 Nature Communications article describing the research.
“By meticulously measuring the composition of ribosomes as cells form different cell types, we found dozens of ribosomal proteins that change in their incorporation into the ribosome. This could lead to hundreds of different ribosome types,” said Genuth, who is now a postdoctoral scholar at UC Berkeley.
“Each of these studies is dragging ribosomes back into the limelight,” Barna said. “Until now, we would not have expected that ribosomes control the development of one of the body’s three germ layers. These are some very unexpected functions for these ancient proteins.”
They’ve created a stir among fellow scientists. “As is often the case in science, Maria’s results were greeted by a gamut of responses and feedbacks,” said leading protein synthesis researcher Nahum Sonenberg, PhD, the Gilman Cheney Chair in Biochemistry at McGill University. “The findings are intriguing, but the general mechanism to explain exactly how the specificity is achieved is yet to be elucidated.”
“All the really cool ideas face controversy,” said NYU’s Schneider. “But this is actually based on a very old idea. Maria has the tools and excellent science background to address whether it’s true, and it very clearly is.”
An international conference is planned for fall 2023 to bring together researchers fleshing out the role of ribosome specificity as many other groups are now following this line of research. Barna is no stranger to weathering controversy with the help of staunch colleagues and collaborators.

“I didn’t come from a privileged background,” she said. “My family was very poor and my father didn’t want me to go to college. I was the first person in my family to go to university, and I was really lucky to have some incredible women mentors. It’s been great to have had, at an early stage in my career, strong women to teach me how to be the best scientist I can be.”
Barna’s mentors gave her the courage to pursue what was at first unlikely fodder for a career on the edge of biomedical science. “I had never imaged I’d be challenging the way ribosomes were thought to work in textbooks,” she said. “Now we have a road map for how ribosomes affect stem cell fate and embryonic development, and it’s really exciting.”
Barna’s focus on ribosomes is unlikely to wane soon.
“There’s an old joke that researchers tend to view cells as being made up of only the proteins or molecules they study,” she reflects.
“And it’s true. My lab members joke that the cell is just a sack of ribosomes. But 60% of a cell’s energy is devoted to making and maintaining ribosomes. To imagine they play no role in gene regulation and cell fate is just a little silly.”

